Instructor: Dr. Natalia Tretyakova, Ph.D. «hyperlink
"mailto:[email protected]"»
- 6-3432
PDB reference
correction and design Dr.chem., Ph.D. Aris
Kaksis, Associate Prof.
mailto:[email protected]
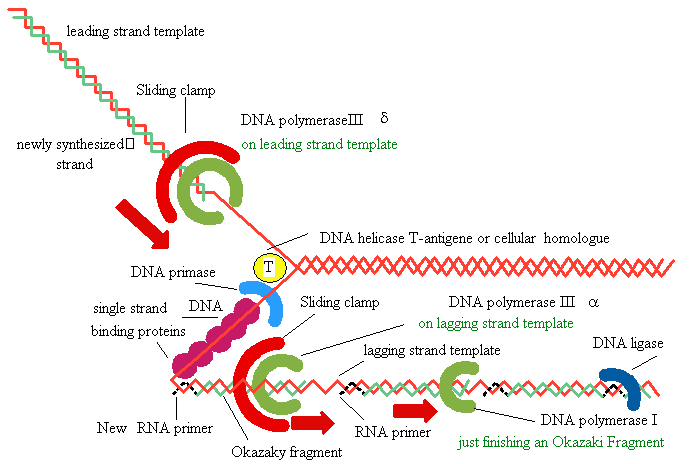
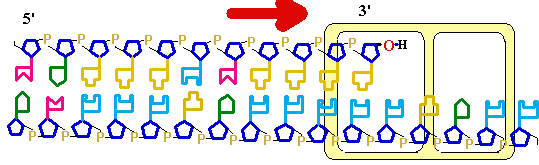
DNA
SynthesisDNA Synthesis
5' 3'
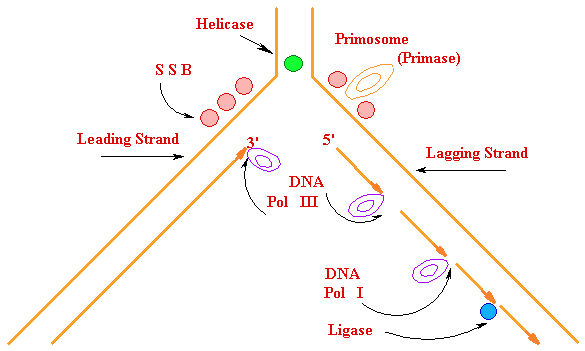
3' 5'
3'
5'
Primer
Synthesis and removal
3'  5'
5'
|| Primase
3'  5'
5'
5'  3'
3'
RNA primer  + || DNA Pol
III
+ || DNA Pol
III
3'  5'
5'
5' 
 --> 3'
--> 3'
|| DNA Pol I
DNA strand
3'
 5'
5'
5'  3'
3'
degraded 






 <-- primer + || DNA
Ligase
<-- primer + || DNA
Ligase
3'  5'
5'
5'  3'
3'
Replication Fork Garland

Fidelity of Polymerization: Absolutely Essential!!
Error Probability =
Polymerization error 10-4
3' --> 5' Nuclease error 10-3
(10-4) •
(10-3)
= 10-7 or 1 in
10,000,000
DNA Synthesis: addition of
new dNTPs follows Watson-Crick rules
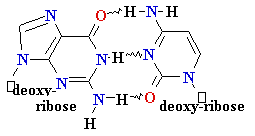
G = C
A=
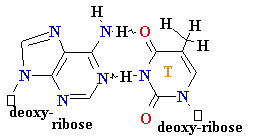
Polymerase
errors
• Very low rate of
misincorporation (1 per 108)
• Errors can occur due to the
presence of minor
tautomers of nucleobases.
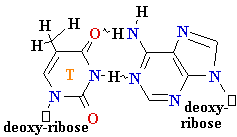
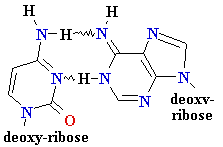
 = A
Cytosine
C = A
Rare tautomer of A
= A
Cytosine
C = A
Rare tautomer of A
Normal base pairing
Mispairing
Proofreading
function of DNA polymerases


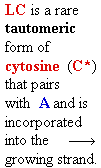
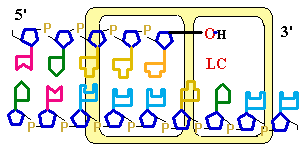

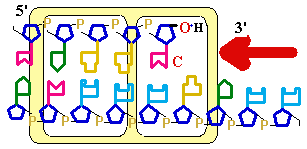
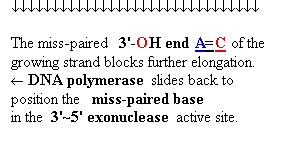
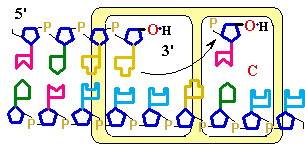
Figure
25-7. An
example of error correction by the 3'--> 5'
exonuclease activity
of DNA polymerase I. Structural analysis has located the exonuclease activity ahead of the polymerase
activity as the enzyme is oriented in its movement
along the DNA. A miss-matched base
(here, a C=A mismatch) impedes
translocation of DNA polymerase I
to the next site. Sliding backward, the enzyme
corrects the mistake with its 5'--> 3' exonuclease
activity, then resumes its polymerase
activity in the 5'--> 3'
direction.
Types
of DNA Mutations
1.
Point mutations:
substitution of one base
pair for another, e.g. A for GC
for GC
•
the most common form of mutation
•
transition; purine
to purine and pyrimidine to pyrimidine
• transversions;
purine to pyrimidine or pyrimidine
to purine
2.
Deletion of one or
more base pairs
3.
Insertion of
one or more base pairs
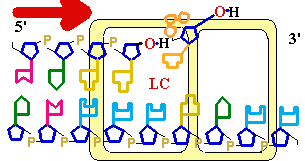
Consider misincorporation due
to a rare tautomer of A
2nd replication -A-
- -
-
1st replication -A(imino)-
-C-
5’-A- -----> -A(imino)
3’- - --> -
- --> - -
||
-
||
-A-
- - Normal replication
- Normal replication
Final result: A --->
G
transition
Mismatch Repair Enzymes
Polymerase I, III error rates: 1 per
107
nucleotides
Observed mutation rate:
1
per 108 - 1 per 1010 nucleotides
Polymerase errors
can be corrected after DNA synthesis!
Repair of nucleotide mismatches:
• Recognize parental
DNA strand (correct base) and daughter
strand (incorrect base)
Parental strand is methylated —CH3: metC 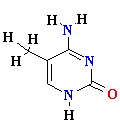 or
or 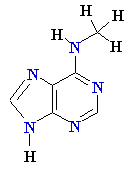 Amet
Amet
2. Replace
a portion of the strand containing erroneous
nucleotide
(between the mismatch and
a nearby methylated
site –up to 1000 nt)
DNA
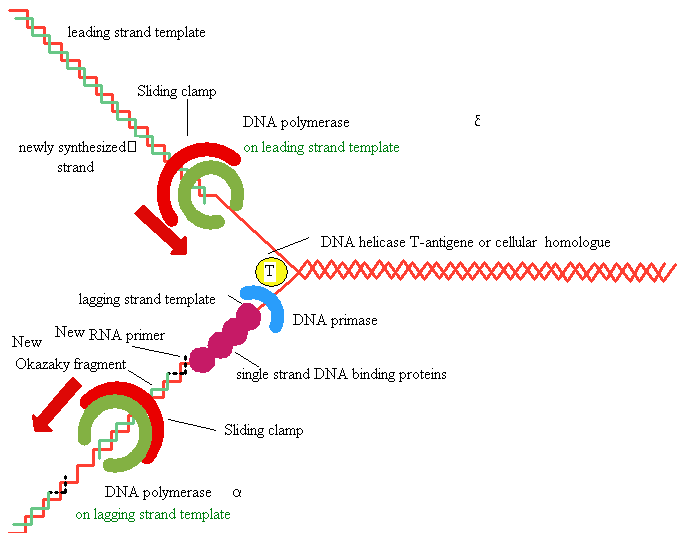
replication in eukaryotes
Several eukaryotic
DNA polymerases are known: alpha, beta, delta, gama -
a
and delta are thought to
be the major chromosomal replicases
Similarities with E.Coli
Always 5’
to 3’ direction -->>
Require a primer
Similarities in active site and tertiary 3° structure
Differences
Eukaryotic replication is
much slower (100
nt/sec)
Several replication
origins
Polymerases are more
specialized (a for lagging strand, d for leading strand)
4. Require special
processing of the chromosomal ends .
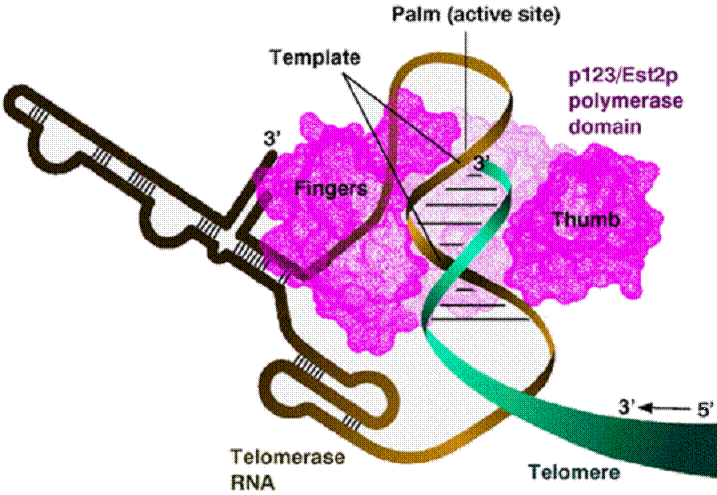
Telomerase preserves
chromosomal ends
• The ends of the linear DNA strand
can not be replicated due to the lack
of a primer
• This would lead to shortening
of DNA strands after
replication
3'  5'
5'
5' 
 3'
<----RNA
primer
3'
<----RNA
primer
• Solution: the chromosomal
ends are extended by DNA telomerase
This enzyme adds hundreds 200÷900
of tandem repeats of a hexa-nucleotide (AGGG
 in humans)
in humans)
to
the parental strand:
3'  5' AGGG
5' AGGG
 AGGG
AGGG
 AGGG
AGGG
 <--- telomere
<--- telomere
5' 

 3'
¯¯¯¯¯¯¯¯
3'
¯¯¯¯¯¯¯¯
3'  5' AGGG
5' AGGG
 AGGG
AGGG
 AGGG
AGGG
 <--- telomere
<--- telomere
5'  3'
3'  CCCAA
CCCAA CCCAA
CCCAA CCCAA<--- RNA primer
CCCAA<--- RNA primer
Telomerase is a
ribonucleoprotein that contains an RNA molecule
used as a template for elongation of the 3’ strand
Central dogma
of molecular biology
DNA ||------------->>
RNA ------------------> Proteins --------->>>>> Cellular action
Replication || transcription
translation
------->>>>>>
|| nucleare
DNA
nucleare <== Reverse
transcription
of telomeres
Notable
exception: retroviruses
RNA ||--------->>
DNA --- --->
RNA -----------------> Proteins --------->>>>> Cellular action
Reverse ||--->
transcription --> transcription ||
translation ||
--------->>>>
DNA cytosolic
nucleare
Reverse
transcriptases (RT)
are RNA
directed DNA Pol
Used by RNA viruses (HIV-I
, human immunoblastosis
virus, Rous sarcoma virus) :
1. Make RNA-DNA hybrid (use its own
RNA as a primer)
2. Make ss DNA by exoribonuclease (RNase H)
activity
3. Make ds DNA incorporate
in the
host genome
 CCCAA
CCCAA CCCAA
CCCAA CCCAA RNA
CCCAA RNA
||<-- RT
 CCCAA
CCCAA CCCAA
CCCAA CCCAA RNA
CCCAA RNA
AGGG
 AGGG
AGGG
 AGGG
AGGG
 - DNA hybrid
- DNA hybrid
||<-- Rnase H --------------------------------------->  CCCAA
CCCAA CCCAA
CCCAA CCCAA RNA
CCCAA RNA
||<-- RT
AGGG
 AGGG
AGGG
 AGGG
AGGG
 ss -DNA
ss -DNA
||<-- RT
 CCCAA
CCCAA CCCAA
CCCAA CCCAA
ds
DNA
CCCAA
ds
DNA
AGGG
 AGGG
AGGG
 AGGG
AGGG
 ds -DNA
ds -DNA
Termination of
Polymerization: The Key to Nucleoside Drugs
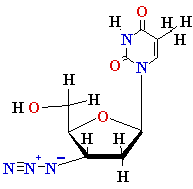
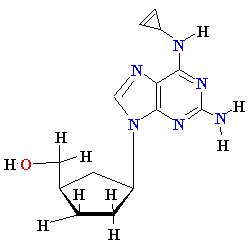
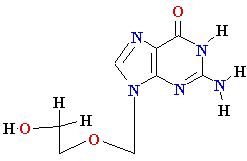
AZT
Ziagen
Acyclovir
Nucleosides
Must Be Converted to Triphosohates to be
Part of DNA and RNA
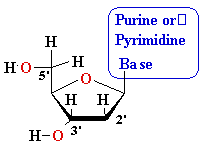
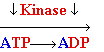
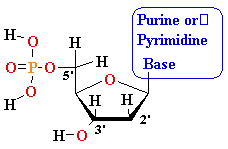

MonoPhosphate
||
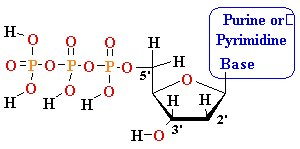

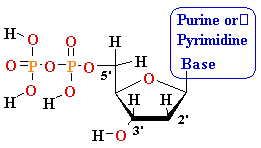
TriPhosphate
DiPhosphate
DNA Damage
Sources of DNA
damage: endogenous
1.
Deamination
2.
Depurination: 10,000/cell/day
3.
Oxidative stress
Sources of DNA
damage: environmental
1. Alkylating agents (drugs, pollutants)
2. X-ray and UV irradiation
3. Diet
4. Smoking
DNA
Damage: oxidative stress
Reactive
oxygen species: HO•, H2O2, 1O2, LOOH

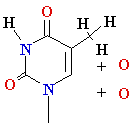
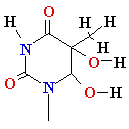
 glycol
glycol
Guanine 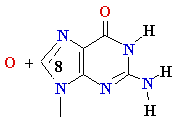
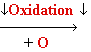
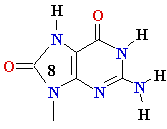 8-oxo-Guanine
8-oxo-Guanine
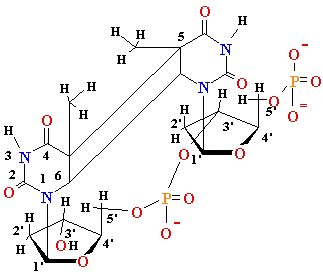
DNA Damage: UV
light
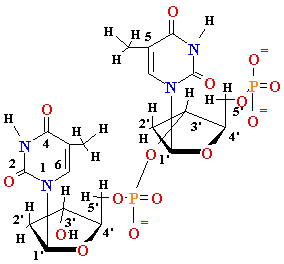
Chemical
Mutagens
Mutations can occur when the normal
bases that are incorporated are changed.
1.
Base analogs or bases that have altered hydrogen bonding capabilities
can cause
transitions.
Ex. Bromouracil and Guanine or 2-aminopurine
and cytosine
2.
Bases can be modified on the DNA by mutagens.
•adenine is
oxidatively deaminated to hypoxanthine, Cytosine
to  , Guanine
to Xanthine
, Guanine
to Xanthine
3. Intercalating Agents
•
insertion and deletion mutants
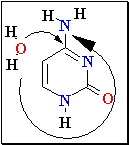
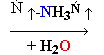
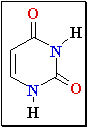 Cytosine Deamination
Cytosine Deamination
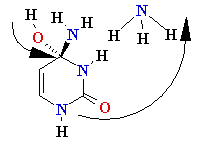
Normal base pairing in DNA
and
an example of mispairing via chemically modified nucleobase
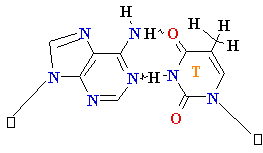
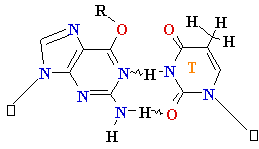
Adenine A=
 O6-Alkyl-Guuanine
O6-Alkyl-Guuanine 
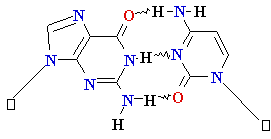
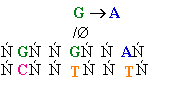
Guanine
G = C Cytosine
Importance of
DNA Repair
• DNA
is the only biological
macromolecule that is repaired. All
others are replaced.
• More than 100 genes are required for DNA repair, even in organisms with
very small genomes.
• Cancer is a consequence of inadequate
DNA repair.
Excision Repair
Takes
advantage of the double-stranded (double
information) nature of the DNA
molecule.
* Mismatch repair
* Base excision repair
* Nucleotide excision repair
Nucleotide
Excision Repair
• Extremely flexible
• Corrects
any damage that both distorts the DNA
molecule and
alters
the chemistry of the DNA molecule.
Nucleotide
Excision Repair
•
In all organisms, NER involves the
following steps:
1. Damage
recognition
2. Binding
of a multi-protein complex at the damaged
site
3. Double
incision of the damaged strand several
nucleotides away
from
the damaged site, on both the 5' and 3' sides
4. Removal of the damage-containing oligonucleotide
from between the two 2 nicks
5. Filling
in of the resulting gap by a DNA
polymerase
6. Ligation
DNA Synthesis:
Take Home Message
1) DNA synthesis is
carried out by DNA polymerases with high
fidelity.
2) DNA
synthesis is characterized by initiation,
priming and processive synthesis
steps and proceeds in 5’--> 3’ direction.
3) Modifications
of DNA base pairs, if not repaired,
can lead to
mutations of the DNA sequence.














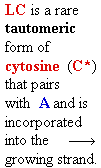



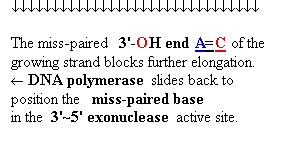


 or
or  Amet
Amet











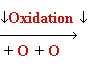


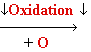
 8-oxo-Guanine
8-oxo-Guanine

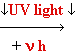


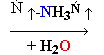
 Cytosine Deamination
Cytosine Deamination