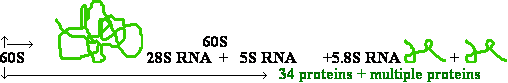
| Eukaryotes |
Prokaryotes |
| 60S subunit |
50 S |
| 40S subunit |
30S |
| 28S RNA 5S RNA 5.8S RNA 18S RNA |
23S RNA 5S RNA - 16S RNA |
| 1st position | 2nd position |
3rd position |
|||
| (5' end) ® |
U |
C |
A |
G |
(3' end) ® |
| U | Phe Phe Leu Leu |
Ser Ser Ser Ser |
Tyr Tyr STOP STOP |
Cys Cys STOP SelenoCys Trp Mitochondria Trp |
U C A G |
| C | Leu Leu Leu Leu |
Pro Pro Pro Pro |
His His Gln Gln |
Arg Arg Arg Arg |
U C A G |
| A | Ile Ile Ile Met init |
Thr Thr Thr Thr |
Asn Asn |
Ser Ser Arg Arg |
U C A G |
| G | Val Val Val Val |
|
Asp Asp Glu Glu |
Gly Gly Gly Gly |
U C A G |
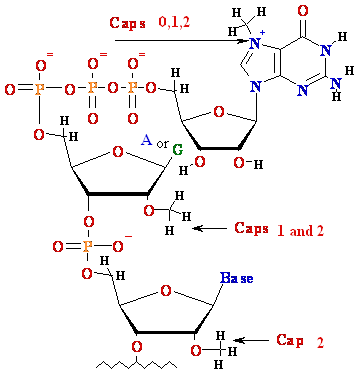 Eukaryotic
mRNA is 5’-Capped
Eukaryotic
mRNA is 5’-Capped
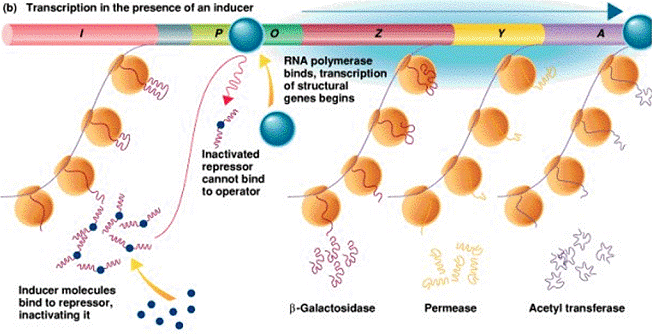
| P |
Promoter |
Binding
site for RNA Pol |
| O |
Operator |
DNA control
region that prevents transcription when repressor is bound |
| I ( R ) |
Regulator repressor gene |
Codes for the repressor
protein |
| Z , Y , A |
Structural genes |
Code for b-galactosidase, galactoside permease, and thiogalactoside transacetylase,
respectively |
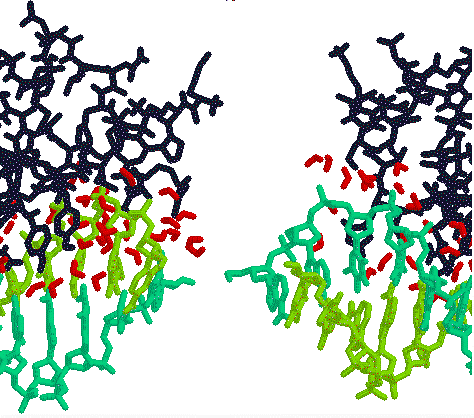
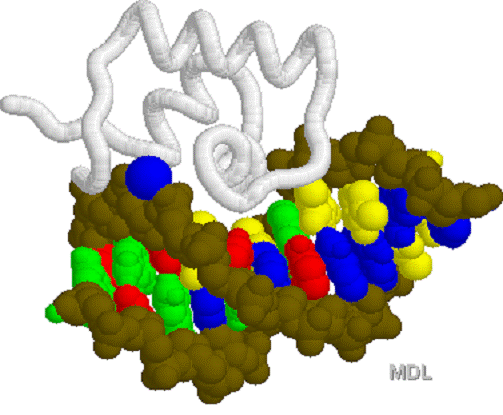 LAC
repressor 1LCC.PDB
LAC
repressor 1LCC.PDB 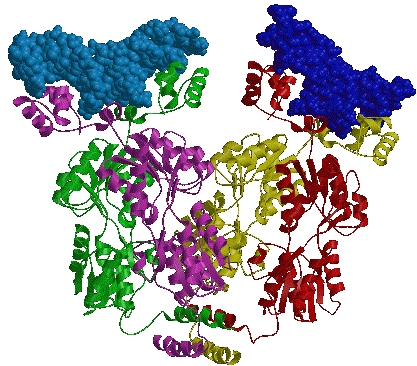
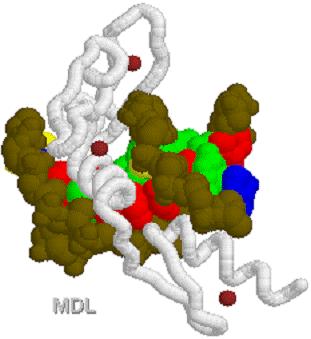

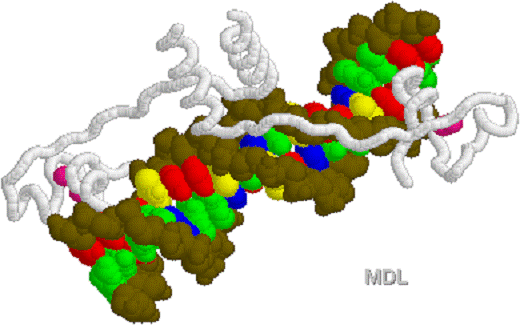 1D66.PDB-Zn2+-Gal4
1D66.PDB-Zn2+-Gal4
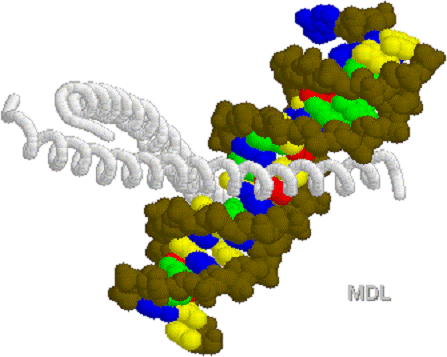


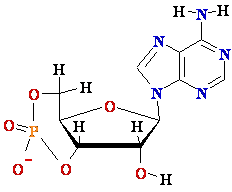 cAMP Promoter sequences for
different genes
cAMP Promoter sequences for
different genes